CandActCFTR
Curated database of candidate therapeutics for the activation of CFTR-mediated ion conductance
Aims of this collaborative project
Cystic fibrosis (CF) is a genetic disease, caused by CFTR which encodes a chloride and bicarbonate transporter expressed in exocrine epithelia throughout the body.
CandActCFTR is a curated compound database which annotates the chemical structure library with information on where and how in the protein life cycle a compound likely interacts, thus comprising a good starting point for modelling the disease and enhancing ligand based approaches (e.g. via eu-openscreen).
Our application CandActBase, adaptable also to other use cases than CFTR compound analysis, and the CandActCFTR data content is provided at https://gitlab.gwdg.de/mnieter1/CandActBase.
In the current extension of CandActCFTR, this ligand-based approach will be complemented by structure-based annotations, including the means to predict the interactions between CandActCFTR substances and CFTR by using existing molecular dynamics trajectories, and by adding more organisation and annotation modules. This approach will help in ranking putative therapeutic substances according to their potential to bind CFTR. Moreover, it will be further extended by integrating results from high-throughput screens from collaborators, helping to rank putative therapeutic substances.
Furthermore CandActCFTR will use public gene expression data to assess transcriptome profiles of CandActCFTR substances and compare differentially expressed genes to gene sets with known relevance for CFTR function, helping to rank putative therapeutic substances according to their potential to modify the cellular transcriptome in favor of CFTR function via Göttingen institutes curated TRANScription FACtor database - TRANSFAC.
Funded by
Deutsche Forschungsgemeinschaft (DFG) - Projekt number 315063128
Currently attached thesis projects
Cystic Fibrosis as a model use case for implementing cell based disease models in systems medicine
- PhD Thesis Liza Vinhoven (finished 11.2022)
Original Papers
Vinhoven L.; Stanke F.; Hafkemeyer S.; Nietert M.: Complementary Dual Approach for In Silico Target Identification of Potential Pharmaceutical Compounds in Cystic Fibrosis. International Journal of Molecular Sciences, Section Biochemistry, Special Issue: Small Molecule Drug Design and Research (accepted October 2022). https://www.mdpi.com/1422-0067/23/20/12351
Voskamp M.; Vinhoven L.; Stanke F.; Hafkemeyer S.; Nietert M.: Integrating Text Mining into the Curation of Disease Maps. Biomolecules 12, no. 9 (September 2022): 1278. https://doi.org/10.3390/biom12091278.
Nietert M.; Vinhoven L.; Auer F.; Hafkemeyer S.; Stanke F.: Comprehensive analysis of chemical structures that have been tested as CFTR activating substances in a publicly available database CandActCFTR
Frontiers in Pharmacology,2021. https://www.frontiersin.org/articles/10.3389/fphar.2021.689205; accepted Nov 2021
Vinhoven L.; Voskamp M.; Nietert M.M. Mapping Compound Databases to Disease Maps—A MINERVA Plugin for CandActBase.
J. Pers. Med., 2021, 11, 1072. https://doi.org/10.3390/jpm11111072; accepted Oct 2021
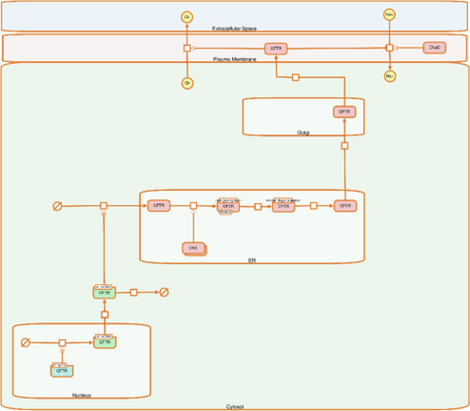
Vinhoven, L.; Stanke, F.; Hafkemeyer, S.; Nietert, M.M. CFTR Lifecycle Map—A Systems Medicine Model of CFTR Maturation to Predict Possible Active Compound Combinations.Int. J. Mol. Sci. 2021, 22(14), 7590; https://doi.org/10.3390/ijms22147590; accepted Jul 2021
Using text mining to annotate and construct biological disease maps - Topics modelling
- Master Thesis Malte Voskamp (finished 12.2021)
- Automated Paper Information retrival and model annotation - Testing if using topics modelling for text sources can help curate a data base like CandActCFTR
- Testing how similar the flags can be derived from the literature compared to manual gold standard
- Automatically extracting topics from the paper stacks and using this to annotate the data
- Providing links to the source positions - Text mining - to extend the annotation network
Project partners
Department of Medical Bioinformatics (UMG) - Group of Chemoinformatics and Imaging
Dr. Manuel Nietert
Medizinische Hochschule Hannover (MHH)
Zentrum für Kinderheilkunde und Jugendmedizin
Klinik für Pädiatrische Pneumologie, Allergologie und Neonatologie
Privatdozentin Dr. Frauke Stanke
Mukoviszidose Institut gGmbH
Dr. Sylvia Hafkemeyer
Press releases
Press release from the DFG:
https://www.dfg.de/foerderung/info_wissenschaft/2022/info_wissenschaft_22_76/index.html (2022) Abschlussveranstaltung der TMF-Projekte
Press release from the Mukoviszidose Institut – gemeinnützige Gesellschaft für Forschung und Therapieentwicklung mbH:
https://idw-online.de/de/news787629 (2022) update results of second funding
https://idw-online.de/de/news726714 (2019) second funding
https://idw-online.de/de/news726714 (2016) first funding
That might also interest you
