Chemoinformatics and Imaging
Background
Our work group aims at providing IT based solutions for biochemical aspects of the systems biology and systems medicine projects.
We develop and adapt software solutions to provide the required tools for medical research.
We accomplish this by applying and extending bioinformatics, cheminformatics and image processing methods to complement systems biology research questions integrating the means for statistical analysis. While extending our methodology toolbox we explicitly try to strengthen the planed analysis by integrating early on the requirements for the statistical analysis of the output of our pipelines.

As more and more image based research is conducted and the number of created images per project increases, we develop the solutions for an automated analysis, e.g. AutoBuSTeD project, but if necessary can even venture out to help improve the acquisition setup. Our newest project Stracyfic is aiming to bring the diagnostic test to the clinical routine by developing the hardware and software for this diagnostic test.
Research foci
For the past years we focus our research in the field of cystic fibrosis, where we still see an unmet demand for IT solutions to help this field.
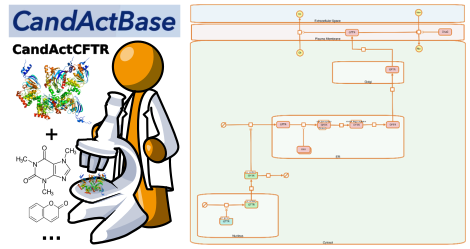
Our CandActCFTR project aims at providing means to distil and curate compound libraries from literature and other online sources and combine this structured information for analysis.
CF is a good target for a system medicine approach
At a first glance, CF is a monocausal disease in which over 2000 putative mutations leading to various forms of phenotypes have been identified. Among these, about 300 variants define the more common types. The CFTR protein is only effective as an integral membrane protein, and as such, it is affected by transcription, translation, folding and degradation, as well as protein traficking processes. Thus, this monogenic disease has multiple sites for potential drug intervention during its life cycle and covers also protein structure variants. This makes it an interesting target for a system medicine approach. The life cycle of the protein offers also multiple modes to obtain information to annotate the system and the existing literature offers various annotations for specific combinations of mutations and read-outs (e.g. protein expression, functional patch clamp measurements, up to structure models and molecular dynamic simulations).
Our newest additon - the CF-Map project - is a MINERVA-based implementation of the CFTR-lifecyle, which provides an interactive view of the underlying systems biology model. It is also the site where we showcase the integration of a specfic compound database like CandActCFTR with a systems biology model and will be extended with more modules in the future.
Web resources
- CandActCFTR a CFTR protein specific curated compound data base
- CF-disease map an interactive systems medicine model, with a focus on CFTR protein maturation
Current projects
Current thesis offers
In the context of the project CandActCFTR work is possible at all levels (internship, bachelor and master). Possible works could be:
1. CandActBase MIRIAM Module - Minimal Information Required in the Annontation of Models
- Implementation of the automated storage and retrieval of Systems Biology Markup Language SBML models - storage, retrieval, updating/extension of annotations and merging of models
- working on Interactive visualization for Systems Model depictions
- conducting comparative statistics analysis
2. CandActBase Protein DataBank/ Molecular Dynamics Trajectory Module
- Automated Target structure model annotation - includes work on storage, retrieval updating/ extension of annotations in the software context of CandAct
- Visualization - e.g. for preparation for virtual docking and linked storage of results, binding site visualization, comparative statistics using similarity descriptions. Developing search modules interfaces and report views.
3. CandActBase SystemsBiology Curation and Annotation Helper Module(s)
3.1 Chemical Information retrieval from PDFs
- Extracting all individual chemical names from the papers and if possible also if available the chemical structures (molecular graphs) from images - integration of existing libraries like OSRA: Optical Structure Recognition
- Automating lookup of structures from chemical names
In the context of the image analysis projects from Manuel Nietert's group, work is possible at all levels (internship, bachelor and master). Possible works could be:
4. Imaging related (possible data sources: e.g. CT, MRI, microscopy, ultrasonic)
- Image Analysis, e.g. detection of objects, quantification, tracing, fusion
- 3D reconstruction and visualization, e.g. from laser scanning microscopy
- developing pipelines to be used later by non-experts: includes GUI development
e.g.
4.1 Automated Lung volume tracing in real time MRI time series diagnosing Pompe
- visualization of results via data fusion and augmentation: geometry, vector and matrix math
- 3D reconstruction
- optional deep learning applied for optimized organ detection / segmentation
- quantification and statistics
- pipeline and GUI
4.2 QuantOS – Quantification of Organoid Swelling in Cystic Fibrosis – with University Verona, Italy
- detection of organoids in microscopy image time series
- quantification and statistics
- pipeline and GUI
4.3 QuantOG – Quantification of Organoid Growth in Cancer Models - with UMG
- detection of organoids in microscopy image time series
- quantification and statistics
- pipeline and GUI
4.4 3D reconstruction from a StereoCameraSystem and overlay with CT data
- visualization of results via data fusion and augmentation: geometry, vector and matrix math
- 3D reconstruction
- detection / segmentation
- quantification and statistics
pipeline and GUI
We have a constant input of new projects from our collaboration partners, sometimes with hardware optimisation tasks as well:
5. Automated optimization of the lighting for a diagnostic test
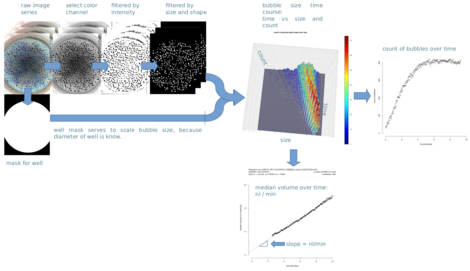
The bubble sweat test to diagnose cystic fibrosis is based on capturing images of the skin and quantifiying the sweat bubble growth behaviour. As the bubbles are detected by their reflection edges in the images optimal lighting is crucial for the robust application. In order to improve the acquisition quality for future measurements we strife to improve this now further.
- establish the control of the ring light by a microcontroller
- integration of image analysis to control the light intensity to optimize the parameter
- geometrical optimization of the setup
- optional: color optimization using different light sources / wave length
Members of the group
Current members of the group
- Dr. Manuel M. Nietert (Head of the group)
- Paul Würzberg (Research Assistant)
Former members of the group
- Dr. Liza Vinhoven, PhD: Cystic Fibrosis as a model use case for implementing cell based disease models in systems medicine(2022)
- Malte Voskamp, Master: Using text mining to annotate and construct biological disease maps (2021)
- David Tschritter, Bachelor: Automatische Quantifizierung von Lysosomenparametern aus Mikroskopiebildern im Kontext der zystischen Fibrose (Hochschule Emden/Leer) (2020)
- Paul Würzburg, Bachelor: Automated Image Analysis by Tracing Object Evolution in Time Series Data for a Diagnostic Test for Cystic Fibrosis (2020)
Open thesis positions
We are currently seeking candidates to join our group at any of the following levels (Bachelor thesis, Master thesis).
Please contact:

contact information
- telephone: +49 551 3961789
- e-mail address: manuel.nietert(at)med.uni-goettingen.de
- location: 2.112
Publications
Pallenberg S. T., Held I., Dopfer C., Minso R., Nietert M.M., Hansen G., Tümmler B., and Dittrich A.M..
Differential Effects of ELX/TEZ/IVA on Organ-Specific CFTR Function in Two Patients with the Rare CFTR Splice Mutations c.273+1G>A and c.165-2A>G.
Frontiers in Pharmacology, Sec. Pharmacology of Ion Channels and Channelopathies, 14 (March 15, 2023). https://doi.org/10.3389/fphar.2023.1153656.Vinhoven L.; Stanke F.; Hafkemeyer S.; Nietert M.:
Complementary Dual Approach for In Silico Target Identification of Potential Pharmaceutical Compounds in Cystic Fibrosis.
International Journal of Molecular Sciences, Section Biochemistry, Special Issue: Small Molecule Drug Design and Research (accepted October 2022). https://www.mdpi.com/1422-0067/23/20/12351Voskamp M.; Vinhoven L.; Stanke F.; Hafkemeyer S.; Nietert M.:
Integrating Text Mining into the Curation of Disease Maps.
Biomolecules 12, no. 9 (September 2022): 1278. https://doi.org/10.3390/biom12091278.Nietert M.; Vinhoven L.; Auer F.; Hafkemeyer S.; Stanke F.:
Comprehensive analysis of chemical structures that have been tested as CFTR activating substances in a publicly available database CandActCFTR
Frontiers in Pharmacology,2021. https://www.frontiersin.org/articles/10.3389/fphar.2021.689205Pallenberg S., T.; Junge S.; Ringshausen C.; F., Sauer-Heilborn A.; Hansen G.; Dittrich A., M.; Tümmler B.; Nietert M.:
CFTR modulation with elexacaftor-tezacaftor-ivacaftor in people with cystic fibrosis assessed by the β-adrenergic sweat rate assay
Journal of Cystic Fibrosis,2021. https://doi.org/10.1016/j.jcf.2021.10.005Vinhoven L.; Voskamp M.; Nietert M.M.
Mapping Compound Databases to Disease Maps—A MINERVA Plugin for CandActBase.
J. Pers. Med. 2021, 11, 1072. doi.org/10.3390/jpm11111072Vinhoven L.; Stanke F.; Hafkemeyer S.; Nietert M.M.
CFTR Lifecycle Map—A Systems Medicine Model of CFTR Maturation to Predict Possible Active Compound Combinations.
Int. J. Mol. Sci. 2021, 22(14), 7590; https://doi.org/10.3390/ijms22147590; accepted Jul 2021.Bleckmann A, Kirchner B, Nietert M, Peeck M, Balkenhol M, Egert D, Rohde TV, Beißbarth T, Pukrop T.
Impact of pre-OP independence in patients with limited brain metastases on long-term survival.
BMC Cancer. 2020 Oct 8;20(1):973. PMID: 33032552 doi.org/10.1186/s12885-020-07459-z.Jo P, Bernhardt M, Nietert M, König A, Azizian A, Schirmer MA, Grade M, Kitz J, Reuter-Jessen K, Ghadimi M, Ströbel P, Schildhaus HU, Gaedcke J.
KRAS mutation status concordance between the primary tumor and the corresponding metastasis in patients with rectal cancer.
PLoS One. 2020 Oct 1;15(10):e0239806. eCollection 2020. PMID: 33002027 doi.org/10.1371/journal.pone.0239806.Uhlig J, Biggemann L, Nietert MM, Beißbarth T, Lotz J, Kim HS, Trojan L, Uhlig A.
Discriminating malignant and benign clinical T1 renal masses on computed tomography: A pragmatic radiomics and machine learning approach.
Medicine (Baltimore). 2020 Apr;99(16):e19725. PMID: 32311963 doi.org/10.1097/MD.0000000000019725.Jo P, Kesruek H, Nietert M, Sahlmann C, Gaedcke J, Ghadimi M, Sperling J.
Inzidenz und Prädiktive Faktoren des Bilateralen Papillären Schilddrüsenkarzinoms.
Zentralblatt für Chirurgie. 2018 Aug;143(4):361-366. Epub 2018 Aug 22, PMID: 30134494 doi.org/10.1055/a-0651-0878.Lowes M, Kleiss M, Lueck R, Detken S, Koenig A, Nietert M, Beissbarth T, Stanek K, Langer C, Ghadimi M, Conradi LC, Homayounfar K.
The utilization of multidisciplinary tumor boards (MDT) in clinical routine: results of a health care research study focusing on patients with metastasized colorectal cancer.
International journal of colorectal disease. 2017; PMID: 28779354, PMCID: PMC5596058, dx.doi.org/10.1007%2Fs00384-017-2871-zLinke F, Harenberg M, Nietert M, Zaunig S, von Bonin F, Arlt A, Szczepanowski M, Weich HA, Lutz S, Dullin C, Janovská P, Krafčíková M, Trantírek L, Ovesná P, Klapper W, Beissbarth T, Alves F, Bryja V, Trümper L, Wilting J, Kube D.
Microenvironmental interactions between endothelial and lymphoma cells: a role for the canonical WNT pathway in Hodgkin lymphoma.
Leukemia. 2017; 31(2):361-372. PMID: 27535218 doi.org/10.1038/leu.2016.232Linke F, Zaunig S, Nietert M, von Bonin F, Lutz S, Dullin C, Janovská P, Beissbarth T, Alves F, Klapper W, Bryja V, Pukrop T, Trümper L, Wilting J, Kube D.
WNT5A: a motility-promoting factor in Hodgkin lymphoma.
Oncogene. 2017; 36(1):13-23. PMID: 27270428 doi.org/10.1038/onc.2016.183Rühlmann F, Nietert M, Sprenger T, Wolff HA, Homayounfar K, Middel P, Bohnenberger H, Beissbarth T, Ghadimi BM, Liersch T, Conradi LC.
The Prognostic Value of Tyrosine Kinase SRC Expression in Locally Advanced Rectal Cancer.
Journal of Cancer. 2017; 8(7):1229-1237. PMID: 28607598, PMCID: PMC5463438 dx.doi.org/10.7150/jca.16980Jo P, Nietert M, Gusky L, Kitz J, Conradi LC, Müller-Dornieden A, Schüler P, Wolff HA, Rüschoff J, Ströbel P, Grade M, Liersch T, Beißbarth T, Ghadimi MB, Sax U, Gaedcke J.
Neoadjuvant Therapy in Rectal Cancer - Biobanking of Preoperative Tumor Biopsies.
Scientifc reports. 2016; 6:35589. PMID: 27752113, PMCID: PMC5067705 doi.org/10.1038/srep35589Styczen H, Nagelmeier I, Beissbarth T, Nietert M, Homayounfar K, Sprenger T, Boczek U, Stanek K, Kitz J, Wolff HA, Ghadimi BM, Middel P, Liersch T, RüschoffJ, Conradi LC.
HER-2 and HER-3 expression in liver metastases of patients with colorectal cancer.
Oncotarget. 2015; 6(17):15065-76. PMID: 25915155, PMCID: PMC4558136 dx.doi.org/10.18632%2Foncotarget.3527von der Heyde S, Wagner S, Czerny A, Nietert M, Ludewig F, Salinas-Riester G, Arlt D, Beißbarth T.
mRNA profling reveals determinants of trastuzumab efciency in HER2-positive breast cancer.
Public Library of Science One. 2015; 10(2):e0117818. PMID: 25710561, PMCID: PMC4339844 dx.doi.org/10.1371%2Fjournal.pone.0117818Conradi LC, Styczen H, Sprenger T, Wolff HA, Rödel C, Nietert M, Homayounfar K, Gaedcke J, Kitz J, Talaulicar R, Becker H, Ghadimi M, Middel P, Beissbarth T, Rüschoff J, Liersch T.
Frequency of HER-2 positivity in rectal cancer and Prognosis.
The American journal of surgical pathology. 2013; 37(4):522-31. PMID: 23282976 doi.org/10.1097/pas.0b013e318272ff4dTanrikulu Y, Nietert M, Scheffer U, Proschak E, Grabowski K, Schneider P, Weidlich M, Karas M, Göbel M, Schneider G.
Scafold hopping by fuzzy pharmacophores and its application to RNA targets.
Chembiochem : a European journal of chemical biology. 2007; 8(16):1932-6. PMID: 17896338 doi.org/10.1002/cbic.200700195Böcker A, Sasse BC, Nietert M, Stark H, Schneider G.
GPCR targeted library design: novel dopamine D3 receptor ligands.
ChemMedChem: Chemistry Enabling Drug Discovery. 2007; 2(7):1000-5. PMID: 17477344 doi.org/10.1002/cmdc.200700067
That might also interest you